Phage display is a pretty cool way of expressing DNA. It’s one of the techniques that make high-throughput screening of protein interactions possible. Another example of high-throughput screening is when your mother gives you a soliloquy of a lecture from the Sun to Pluto and all you heard was she’s going cut your allowance in half – the only thing that matters attention. “Noooooo” That analogy aside, the first thing to note is that the phage is a virus that exclusively infects bacteria. The more formal term for a phage is actually a “bacteriophage”. Phages are commonly employed as DNA vectors – carriers of DNA, be it fragments of human DNA or synthesized DNA in the laboratory – because they can easily be used to express recombinant DNA genes upon infection to typical host bacteria such as E. coli.
Now it is basic knowledge that the DNA of a cell always stays inside the nucleus – it never goes out. That is why during the process of gene expression, the mRNA (messenger RNA) is needed because it does exactly what it’s named for: it’s a messenger of the information inside the nucleus (i.e. the DNA) to the ribosomes outside the nucleus. Because of this limitation of DNA, analyzing or isolating it usually requires an additional step (e.g. destruction of the nuclear membrane, etc.). What’s unique about the phage display technique (as described by George P. Smith & Valery Petrenko back in 1997) is that this limitation is overruled because the DNA inserted into the phage is actually incorporated and expressed as a coat protein. Think of phage display as a way of putting a specific protein (encoded from a fused DNA) on top of the phage’s coat. Yes, it’s literally on the trench coat of our dear phage. Thus, techniques used to analyze free compounds in solution may be used in the analysis of the phage with the expressed protein (and consequently the inserted DNA to the phage) on its coat. Using phage display, a “library” of phages expressing billions of different proteins can be generated. The phage display library can then be used to screen the billions of different proteins for specific interactions with target molecules.
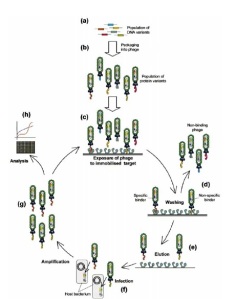
source: Willats, W.G., Phage display: practicalities and prospects. Plant Mol Biol, 2002. 50(6): p. 837-54.
click on image to enlarge
There are multitudes of both current and promising applications of phage display libraries. For the introductory purpose of this article, let’s examine 2 applications. The first one is a general in vitro application of phage display libraries in screening for their selective binding capacities to a target molecule. This is done by first attaching the target molecule on a microtiter plate (i.e lots of copies of the target molecule are immobilized on a plate), followed by the addition of the entire phage display library. The phages are given enough time to bind to the target molecule, after which the plate is washed to take out all non-binding phages. This process is called “biopanning” because of its similarity to the process of panning out gold. The binding phages can then be separated from the target molecules through elution and their DNA may be analyzed by first allowing them to infect a host bacteria and thereby the host bacteria amplifies or increases them in number. Once an ample amount is available, the DNA may then be sequenced and analyzed for its specific interaction with the target molecule. Another application of phage display libraries is an in vivo identification of “homing peptides” to specific organs or tissues in the body. This is usually done by injecting the entire phage display library (recall that this so-called library is actually a population of phages with different proteins expressed on their coats) into the tail vein of a mouse. The phages are then given enough time to travel along the circulatory system and settle to wherever they may end up binding with. The phages that have attached to the organ or tissue of interest are then “rescued” and subsequently analyzed for their DNA using the same method as described earlier. The information gathered from the analysis of these phage display libraries can be used for target drug delivery, identifying specific tumor markers, and development of highly specific antibodies, just to name a few. So overall, a really remarkable technique, don’t you think? :)
[1] Chien, S., C.Y.P. Chen, and Y.C. Fung, An introductory text to bioengineering. Vol. 4. 2008, Singapore: World Scientific.
[2] Willats, W.G., Phage display: practicalities and prospects. Plant Mol Biol, 2002. 50(6): p. 837-54.
[3] Smith, G.P. and V.A. Petrenko, Phage display. Chemical Reviews, 1997. 97(2): p. 391-410.